rVarBase annotates variant's regulatory feature in three fields: chromatin state of the region surrounding variant, regulatory elements overlapped with variant, and variant's potential target genes. It also provides optioned extended annotation for variants, including: LD-proxies of known SNP, SNP/CNV that is overlapped with or located in queried variant, traits (disease and expression quantitative trait) associated with variant. rVarBase is an updated version of the database rSNPBase, it is consistent with the old version in utilizing experimentally supported regulatory elements from ENCODE and other data resources to make relevant annotation (such as involved regulatory manner and potential target gene).rVarBase is different from the old version in several new features:
- New variant types
- New dimension of annotation
- New regulatory manner
- More detailed annotation on variant overlapped TFBS
- More extended data
- New search manner
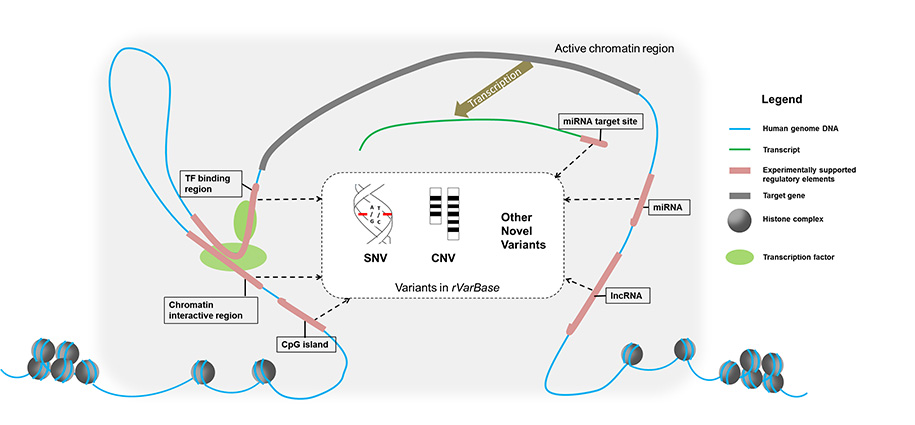
Citation: Guo, L., Du, Y., Qu, S., & Wang, J. (2015). rVarBase: an updated database for regulatory features of human variants. Nucleic acids research, gkv1107 pubmed





